Group Leader 2019-
Habilitation: "Determination of transmembrane protein structure and function considering their evolutionary diversity", Faculty of Chemistry, University of Warsaw.
Białka błonowe stanowią 20-30 % ludzkiego genomu. Stanowią trudny obiekt do badań eksperymentalnych i w związku z tym jest ich znacznie mniej w Protein Data Bank niż białek globularnych. Biologia obliczeniowa stanowi uzupełnienie badań eksperymentalnych a w wielu przypadkach je zastępuje - właśnie w przypadku białek błonowych. Zajmujemy się przewidywaniem struktur białek transbłonowych metodami modelowania porównawczego i de novo, badaniem dynamiki i mechanizmów ich działania oraz projektowaniem dla nich substancji o znaczeniu farmakologicznym oraz ich optymalizacją.
Zapraszamy studentów do realizacji prac dyplomowych i doktorskich. Przykładowe tematy prac licencjackich i magisterskich: "Mechanizmy aktywacji receptora sekretynowego", "Odróżnianie agonistów od antagonistów receptorów GPCR", "Projektowanie nowych substancji czynnych modulujących działanie receptora kalcytoninowego", "Potencjały statystyczne w modelowaniu białek transbłonowych", "Badanie dynamiki kompleksu ligand-receptor na przykładzie receptora glukagonowego", "Aplikacje internetowe w modelowaniu receptorów GPCR", "Badanie mechanizmów transportu przez błonę komórkową na przykładzie rodziny transporterów nukleozydowych".
GPCR receptors constitute large and diverse group of drug targets. This project is aimed at design of self-learning systems that are fitted to specific GPCR pharmacology. Such systems will decrease the number of compounds that need to be tested experimentally in a pre-clinical phase of drug discovery. This project involves ligand-based and structure-based approaches to drug design. As a result, data on the actual receptor response for a ligand together with its physicochemical properties can be combined with the structural description of the ligand-receptor complex.
If you are interested in participation send your CV to dlatek@chem.uw.edu.pl.
Homology modeling of GPCR structures & drug design
Activation switches
De novo structure prediction & pharmacogenomics
SLC28
Model building from multiple templates & sequence profiles comparison
GPCRM, GPCRDOCK 2013, De novo structure prediction, NMR-guided modeling
Dynamics of biological processes on a molecular level
S1P1, FPR1, class B GPCRs, VPAC1, V-ATPase
Structure-based drug design & lead optimization Glucagon receptors, VPAC1, V-ATPase, INPHARMA - NMR
We developed three web applications for modeling of G protein-coupled receptors: GPCRVS for GPCR virtual screening enhanced by machine learning, a previous web service for molecular docking with Autodock Vina against several class B GPCRs - GUT-DOCK and finally GPCRM that allowed us to win the GPCR Dock 2013 competition.
One of our recent projects is a GPCR ligand database with curated data sets for training of machine learning algorithms - Resources for GPCR research
Early programs for using sparse NMR data in protein target structure prediction and drug design are CABS-NMR and INPHARMA (co-developer).
CABS-CONT was the earliest usage of interresidual contacts predicted from, e.g., correlated mutations, MSA or HMMs, in protein folding.
Scripts for processing of MD trajectories.
Molecular dynamics used to observe activation of chemotactic receptors: simulations data for C5aR1 (stable interactions with Gi protein subunits) and C5aR2 (unstable).
GUT-DOCK (molecular docking for class B GPCR receptors i.a. gut hormone receptors) has moved to https://gut-dock.chem.uw.edu.pl
A brief introduction on how to use GPCRVS: GPCRVS tutorial
Habilitation: "Determination of transmembrane protein structure and function considering their evolutionary diversity", Faculty of Chemistry, University of Warsaw.
"Structural studies based on the low sequence similarity of G-protein coupled receptors activated by hormones and allosteric modulators.", National Science Centre in Poland, grant no DEC-2012/07/D/NZ1/04244
Post-doc in the Laboratory of Prof. Filipek in the International Institute of Molecular and Cell Biology in Warsaw. Short-term post-doc fellowships: EMBL-Heidelberg, University of California San Francisco.
PhD studies in the Laboratory of Prof. Kolinski, Faculty of Chemistry, University of Warsaw. PhD thesis title: "Protein structure determination using sparse experimental data", MNiSW grant for PhD students: „Modeling of protein structures based on fragmentary data from simple NMR experiments”.
Supervisor: Prof. A. Kolinski, Faculty of Chemistry, University of Warsaw.
P. Dragan, V. Gratio, T. Voisin, A. Couvineau, D. Latek Non-canonical Gq Activation by Orexin Receptor Type 2 and Lemborexant Observed in Microsecond Molecular Dynamics Simulations. Scientific Reports, accepted.
D. Latek, K. Prajapati, P. Dragan, M. Merski, P. Osial GPCRVS - AI-driven Decision Support System for GPCR Virtual Screening. Int. J. Mol. Sci. 2025, 26(5), 2160. https://doi.org/10.3390/ijms26052160
D. Aranda-Garcia, T. M. Stepniewski, M. Torrens-Fontanals, A. Garcia-Recio, M. Lopez-Balastegui, B. Medel-Lacruz, A. Morales-Pastor, A. Peralta-Garcia, M. Dieguez-Eceolaza, D. Sotillo-Nunez, T. Ding, M. Drabek, C. Jacquemard, J. Jakowiecki, W. Jesper, M. Jimenez-Roses, V. Jun-Yu-Lim, A. Nicoli, U. Orzel, A. Shahraki, J. K. S. Tiemann, V. Ledesma-Martin, F. Nerin-Fonz, S. Suares-Dou, O. Canal, G. Pandy-Szekeres, J. Mao, D. E. Gloriam, E. Kellenberger, D. Latek, R. Guixa-Gonzales, H. Gutierrez-de-Teran, I. G. Tikhonova, P. W. Hildebrand, M. Filizola, M. Madan Babu, A. Di Pizio, S. Filipek, P. Kolb, A. Cordomi, T. Giorgino, M. Marti-Solano, J. Selent Large scale investigation of GPCR molecular dynamics data uncovers allosteric sites and lateral gateways. Nature Communications 16, 2020 (2025). https://doi.org/10.1038/s41467-025-57034-y
Gratio V., Dragan P., Garcia L., Saveanu L., Nicole P., Voisin T., Latek D., Couvineau A. Pharmacodynamics of the orexin receptor type 1 (OX1R) in colon cancer cell models: A two-sided nature of antagonistic ligands resulting from partial dissociation of Gq. British Journal of Pharmacology 2024, 182 (7), 1528-1545 https://doi.org/10.1111/bph.17422
D. Latek, K. Prajapati, P. Dragan, M. Merski, P. Osial GPCRVS - AI-driven decision support system for GPCR virtual screening. bioRxiv, 2024.10. 28.620626 https://doi.org/10.1101/2024.10.28.620626
Dragan P., Latek D. The two-sided impact of beta-adrenergic receptor ligands on inflammation. Current Opinion in Physiology, 2024, 41, 100779. https://doi.org/10.1016/j.cophys.2024.100779
Sanmukh, S.G., Krzykawska-Serda, M., Dragan, P., Baron, S., Lobaccaro, JM.A., Latek, D. Is Cancer Our Equal or Our Better? Artificial Intelligence in Cancer Drug Discovery. In: Interdisciplinary Cancer Research. Springer, Cham. (2024) https://doi.org/10.1007/16833_2024_326 The chapter is available upon request from authors.
P. Dragan, K. Joshi, A. Atzei, D. Latek Keras/TensorFlow in Drug Design for Immunity Disorders. Int. J. Mol. Sci. 2023, 24, 15009. https://doi.org/10.3390/ijms241915009
P. Dragan, M. Merski, S. Wisniewski, S.G. Sanmukh, D. Latek Chemokine Receptors - Structure-Based Virtual Screening Assisted by Machine Learning. Pharmaceutics 2023, 15(2), 516; https://doi.org/10.3390/pharmaceutics15020516
SG Sanmukh, N Jose dos Santos, C Nascimento Barquilha, M de Carvalho, P Pintor dos Reis, FK Delella, HF Carvalho, D Latek, T Feher, SL Felisbino. Oncology Letters 25.2 (2023): 86. https://doi.org/10.3892/ol.2023.13672
Dragan P, Atzei A, Sanmukh SG, Latek D. Computational and experimental approaches to probe GPCR activation and signaling. Prog Mol Biol Transl Sci. 2022;193(1):1-36. https://doi.org/10.1016/bs.pmbts.2022.06.001
S. Wisniewski; P. Dragan; A. Makal; D. Latek Helix 8 in chemotactic receptors of the complement system. PLoS Comput Biol 2022, 18(7):e1009994. https://doi.org/10.1371/journal.pcbi.1009994 Additional data available.
Langer, I.; Jeandriens, J.; Couvineau, A.; Sanmukh, S.; Latek, D. Signal Transduction by VIP and PACAP Receptors. Biomedicines 2022, 10, 406. https://doi.org/10.3390/biomedicines100204061
I. Langer, D. Latek. Drug Repositioning For Allosteric Modulation of VIP and PACAP Receptors. Frontiers in Endocrinology 2021, 12:711906. https://doi.org/10.3389/fendo.2021.711906 in Front. Endocrinol. Sec. Cellular Endocrinology, Vol. 14: 'Peptide-binding GPCRs coming of age' https://doi.org/10.3389/fendo.2023.1189508
M. Mizera, D. Latek. Ligand-receptor interactions and machine learning in GCGR and GLP-1R drug discovery. Int. J. Mol. Sci. 2021, 22(8), 4060.
I. Rodriguez-Espigares et al. GPCRmd uncovers the dynamics of the 3D-GPCRome. Nature Methods 2020, 17, 777-787.
M. Mizera, D. Latek, J. Cielecka-Piontek. Virtual Screening of C. Sativa Constituents for the Identification of Selective Ligands for Cannabinoid Receptor 2. Int. J. Mol. Sci. 2020, 21(15), 5308.
D Latek*, I Langer, KA Krzysko, L Charzewski. A Molecular Dynamics Study of Vasoactive Intestinal Peptide Receptor 1 and the Basis of Its Therapeutic Antagonism. Int. J. Mol. Sci. 2019, 20, 4348.
Latek D*, Rutkowska E, Niewieczerzal S, Cielecka-Piontek J. Drug-induced diabetes type 2: In silico study involving class B GPCRs. PLoS ONE 2019 14(1): e0208892. https://doi.org/10.1371/journal.pone.0208892
Miszta P, Pasznik P, Jakowiecki J, Sztyler A, Latek D, Filipek S*. GPCRM: a homology modeling web service with triple membrane-fitted quality assessment of GPCR models. Nucleic Acids Res. 2018;46(W1):W387‐W395. doi:10.1093/nar/gky429
Latek D*, Bajda M, Filipek S*. A Hybrid Approach to Structure and Function Modeling of G Protein-Coupled Receptors, J Chem Inf Model 2016, 56(4), 630-641, DOI: 10.1021/acs.jcim.5b00451
Yuan S, Wu R, Latek D, Trzaskowski B, Filipek S*. Lipid Receptor S1P1 Activation Scheme Concluded from Microsecond All-Atom Molecular Dynamics Simulations. PLoS Comput Biol 2013, 9(10): e1003261. doi:10.1371/journal.pcbi.1003261.
Latek D*, Pasznik P, Carlomagno T & Filipek S*. Towards Improved Quality of GPCR Models by Usage of Multiple Templates and Profile-Profile Comparison. PLOS ONE 2013, 8, e56742, doi: 10.1371/journal.pone.0056742.
Yuan S, Ghoshdastider U, Trzaskowski B, Latek D, Debinski A, Pulawski W, Wu R, Gerke V & Filipek S*. The role of water in activation mechanism of human N-formyl peptide receptor 1 (FPR1) based on molecular dynamics simulations. PLOS ONE 2012, 7, e47114, doi: 10.1371/journal.pone.0047114.
Latek D, Kolinski M, Ghoshdastider U, Debinski A, Bombolewski R, Plazinska A, Jozwiak K & Filipek S*. Modeling of ligand binding to G protein coupled receptors: cannabinoid CB1, CB2 and adrenergic beta 2 AR. J Mol Model 2011, 17, 2353-2366, doi:10.1007/s00894-011-0986-7.
Latek D, Modzelewska A, Trzaskowski B, Palczewski K & Filipek S*. G protein-coupled receptors--recent advances. Acta Biochim Pol 2012, 59, 515-529.
Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A & Filipek S*. Action of molecular switches in GPCRs--theoretical and experimental studies. Curr Med Chem 2012, 19, 1090-1109.
Latek D. Rosetta Broker for membrane protein structure prediction: concentrative nucleoside transporter 3 and corticotropin-releasing factor receptor 1 test cases, BMC Structural Biology 2017, 17:8, DOI: 10.1186/s12900-017-0078-8.
Dreisigacker S, Latek D, Bockelmann S, Huss M, Wieczorek H, Filipek S, Gohlke H, Menche D & Carlomagno T*. Understanding the inhibitory effect of highly potent and selective archazolides binding to the vacuolar ATPase. J Chem Inf Model 2012, 52, 2265-2272, doi: 10.1021/ci300242d.
Latek D* & Kolinski A. CABS-NMR--De novo tool for rapid global fold determination from chemical shifts, residual dipolar couplings and sparse methyl-methyl NOEs. J Comput Chem 2011, 32, 536-544, doi: 10.1002/jcc.21640.
Latek D*, Ekonomiuk D & Kolinski A. Protein structure prediction: combining de novo modeling with sparse experimental data. J Comput Chem 2007, 28, 1668-1676, doi: 10.1002/jcc.20657.
Codutti L, Skjaerven L, Angelini A, Grimaldi M, Latek D, Monecke P, Dreyer M & Carlomagno T*. Accounting for conformational variability in protein-ligand docking with NMR-guided rescoring. J Am Chem Soc 2013, 135(15), 5819-27.
Sanmukh, S.G., Krzykawska-Serda, M., Dragan, P., Baron, S., Lobaccaro, JM.A., Latek, D. Is Cancer Our Equal or Our Better? Artificial Intelligence in Cancer Drug Discovery. In: Interdisciplinary Cancer Research. Springer, Cham. (2024) https://doi.org/10.1007/16833_2024_326
Dragan P, Atzei A, Sanmukh SG, Latek D. Computational and experimental approaches to probe GPCR activation and signaling. Prog Mol Biol Transl Sci. 2022;193(1):1-36. doi:10.1016/bs.pmbts.2022.06.001.
D Latek, B Trzaskowski, S Niewieczerzał, P Miszta, K Młynarczyk, A Dębiński, W Puławski, S Yuan, A Sztyler, U Orzeł, J Jakowiecki, S Filipek*. Modeling of Membrane Proteins. In: Liwo A. (eds) Computational Methods to Study the Structure and Dynamics of Biomolecules and Biomolecular Processes. Springer Series on Bio- and Neurosystems, 2019, vol 8. Springer, Cham
Gront D, Latek D, Kurcinski M & Kolinski A*. (2008) Template-Free Predictions of Three-Dimensional Protein Structures: From First Principles to Knowledge-Based Potentials. In Prediction of Protein Structures, Functions, and Interactions, pp. 117-141. John Wiley & Sons, Ltd.
Kufareva I, Katritch V, Participants of GPCR Dock 2013, Stevens RCS, Abagyan R. (2014) Advances in GPCR Modeling Evaluated by the GPCR Dock 2013 Assessment: Meeting New Challenges. Structure , 22(8) , 1120 – 1139

A new special issue edited by Dorota Latek has been opened for submissions. Understanding the ligand recognition, activation, signaling, and regulation of these receptors is of prime importance not only to decipher aspects of human physiology but also for developing of novel therapeutics.

'For the first time and probably not the last a scientific breakthrough enabled by artificial intelligence (AI) has been recognized with a Nobel prize.' Nobel Prize was awarded to John Jumper and Demis Hassabis for AlphaFold and David Baker for Rosetta.

'With the [presented at Oncornet 2.0] tools and model systems for the oncogenic chemokine receptor CXCR4 (involved in cancer and metastasis) and the atypical receptor ACKR3 in place, the signaling networks and translational effects can be studied in more detail.'
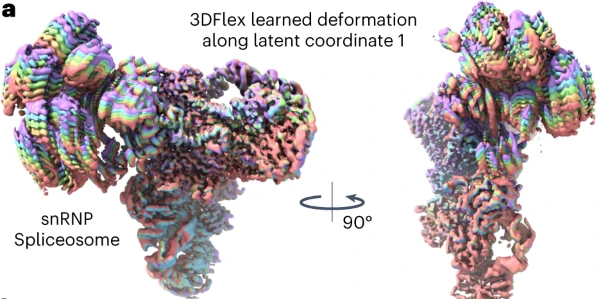
'A deep learning algorithm maps out the continuous conformational changes of flexible protein molecules from single-particle cryo-electron microscopy images, allowing the visualization of the conformational landscape of a protein with improved resolution of its moving parts.'
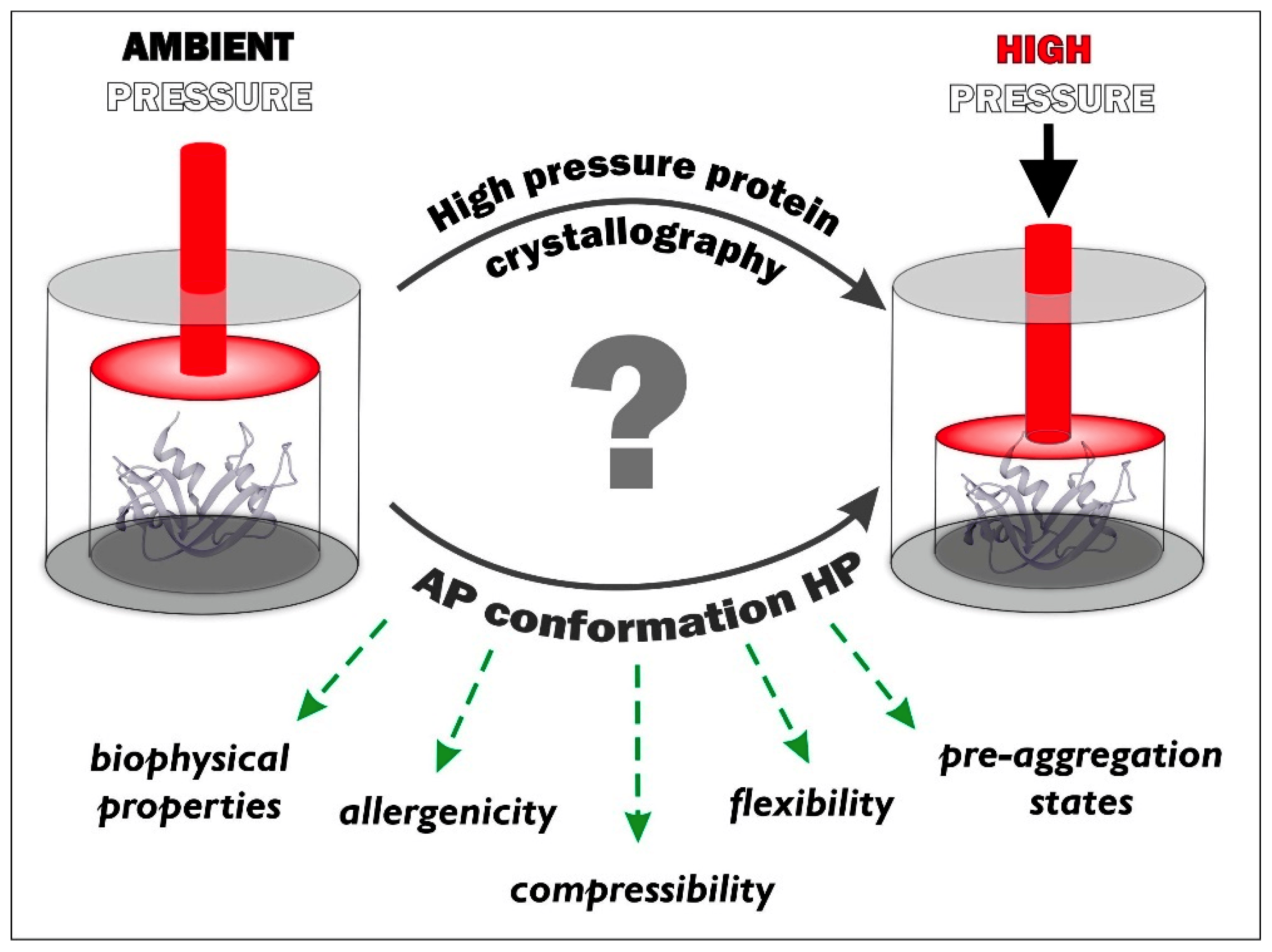
'Since its introduction in the early 1970s, high pressure crystallography (HPX) has shown great potential for the investigation of different types of matter. Using diamond anvil cells, HPX is an emerging technique that has been rapidly implemented, making it available to biologists, and there is immense potential for utilizing this technique in biological systems in the future...'
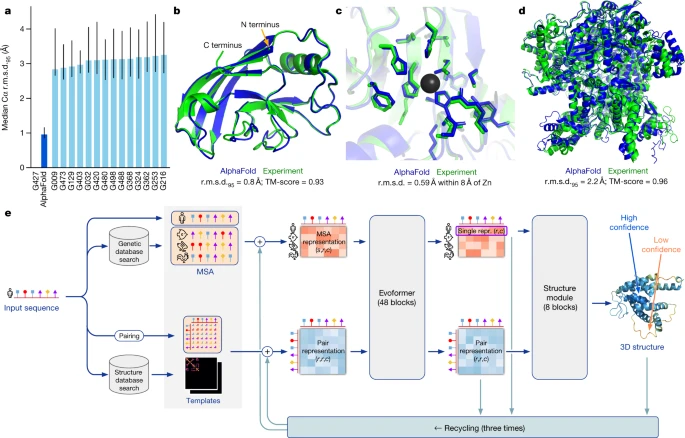
'AlphaFold is a novel machine learning approach that incorporates physical and biological knowledge about protein structure, leveraging multi-sequence alignments, into the design of the deep learning algorithm.'
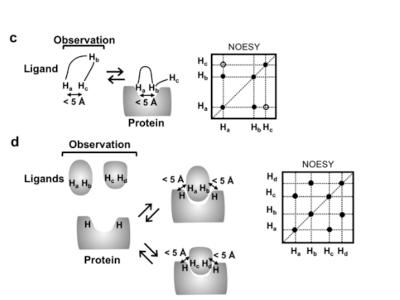
'nuclear magnetic resonance (NMR) spectroscopy in solution is a key tool for analysing function-related conformational equilibria in GPCRs as they relate to allosteric coupling...'
'GPR158, a class C orphan GPCR, functions in cognition, stress-induced mood control, and synaptic development. Among class C GPCRs, GPR158 is unique as it lacks a Venus flytrap-fold ligand-binding domain and terminates Galphai/o protein signaling through the RGS7-Gbeta5 heterodimer. Here, we report the cryo-EM structures of GPR158 alone and in complex with one or two RGS7-Gbeta5 heterodimers.'
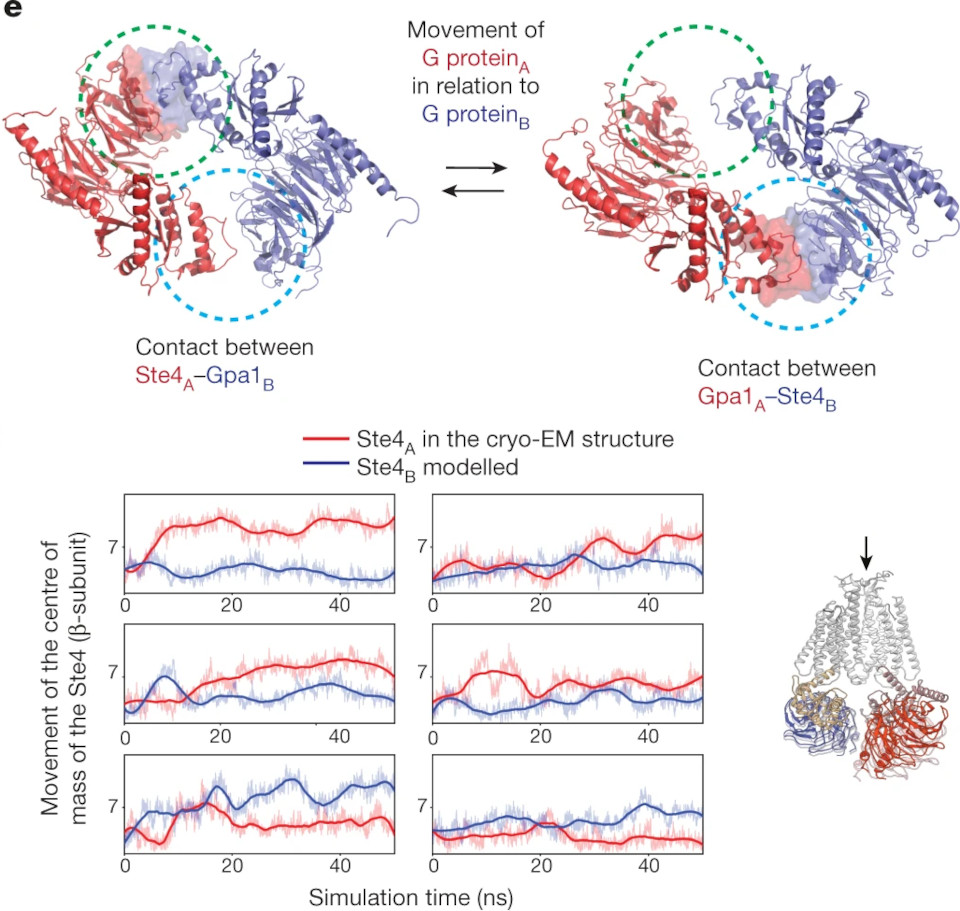
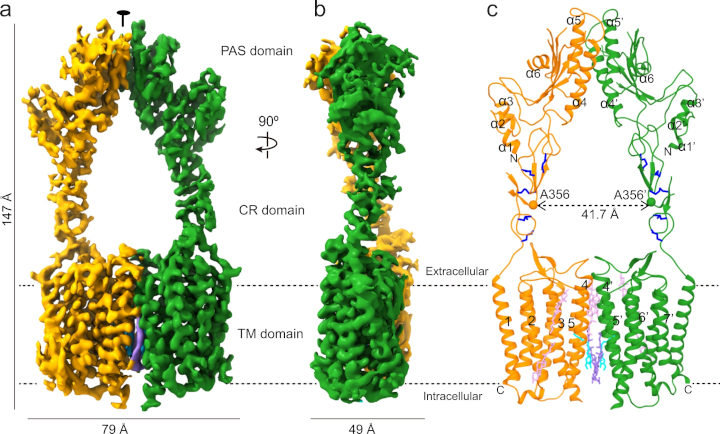
'GPCRs are divided phylogenetically into six classes, denoted A to F. More than 370 structures of vertebrate GPCRs (belonging to classes A, B, C and F) have been determined [...]. Here we determine the structure of a [first] class D GPCR, the Saccharomyces cerevisiae pheromone receptor Ste2, in an active state coupled to the heterotrimeric G protein Gpa1–Ste4–Ste18. Ste2 was purified as a homodimer coupled to two G proteins.'
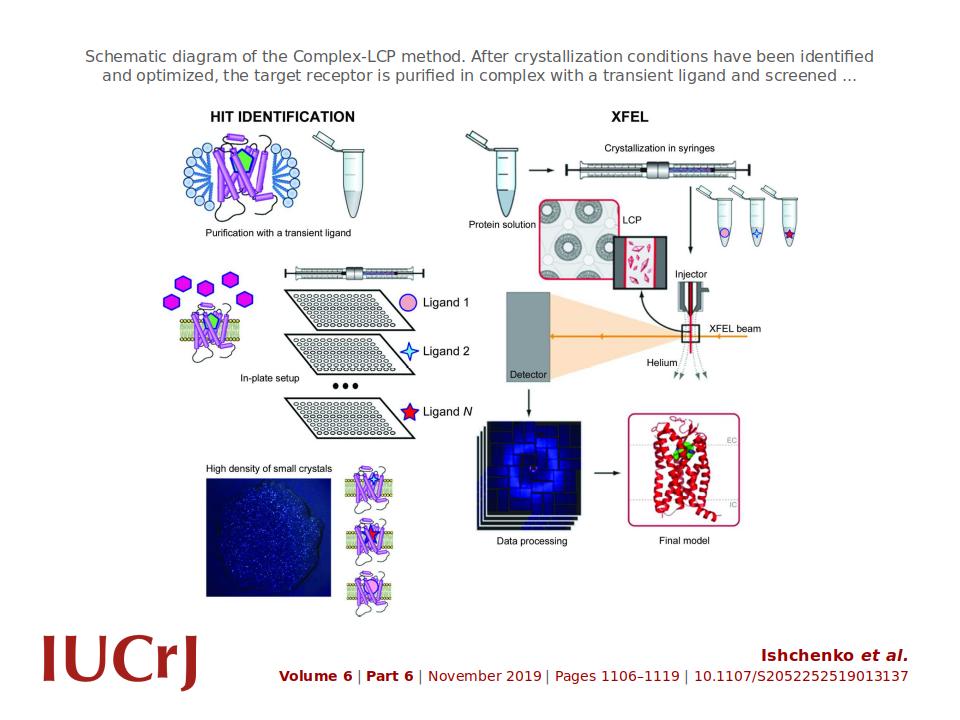
'Rational structure-based drug design (SBDD) relies on the availability of a large number of co-crystal structures to map the ligand-binding pocket of the target protein and use this information for lead-compound optimization via an iterative process.[...] Here, a method is presented for the rapid determination of multiple co-crystal structures for a target GPCR in complex with various ligands, taking advantage of the serial femtosecond crystallography approach, which obviates the need for large crystals and requires only submilligram quantities of purified protein.'
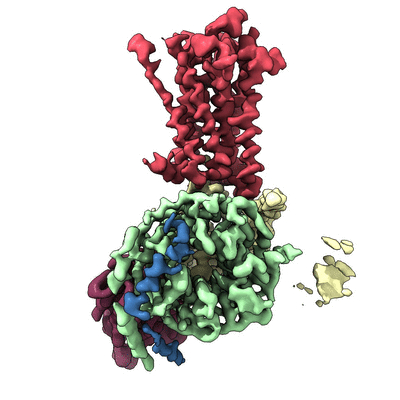
'Classically, G-protein-coupled receptors (GPCRs) are thought to activate G protein from the plasma membrane and are subsequently desensitized by Beta-arrestin (Beta-arr). However, some GPCRs continue to signal through G protein from internalized compartments, mediated by a GPCR–G protein–Beta-arr ‘megaplex’. [...] Here, we present its cryo-electron microscopy structure'
'The world’s first superconducting megahertz repetition rate hard X-ray free-electron laser (XFEL), the European XFEL, began operation in 2017, featuring a unique pulse train structure with 886 ns between pulses. [...] Here, we report the first membrane protein megahertz SFX experiment, where we determined a 2.9 Å-resolution SFX structure of the large membrane protein complex, Photosystem I, a > 1 MDa complex containing 36 protein subunits and 381 cofactors.'